Human cancer genome sequencing studies have generated ample, publicly available data, and analyses of these data have substantially broadened the knowledge of somatic mutations accumulating in tumours. Analysis of mutational signatures focuses on how characteristic somatic DNA mutation patterns reflect the contributions of particular mutagenic processes to cancer development, and it is thus of key importance for cancer etiology and carcinogen exposure studies. A considerable fraction of the compendium of mutational signatures extracted from cancer genome sequencing studies remains without etiological explanation, and mutational signatures alone may be insufficient in explaining cancer causes. These observations highlight the need for innovative and integrated experimental strategies, models, and computational approaches.
The MUTSPEC 2.0 project was developed to accommodate a highly integrated design to identify mutational (and other toxicogenomic) signatures of carcinogens derived by genome-scale sequencing analysis of mutually complementary and cross-validating systems. These include in vitro and in vivo exposure models, suitable collections of human cancer and non-cancer tissues, and data from large-scale public repositories (see the figure). By applying this integrated research strategy, the MUTSPEC 2.0 project delineates signatures of various candidate carcinogens, including food contaminants (e.g. mycotoxins), dietary compounds, components of medicinal products, chemicals in tobacco smoke, and various environmental contaminants. The toxicogenomic signatures from analysed samples are subsequently linked to -omics patterns found in the human cancer genomics databases. The Epigenomics and Mechanisms Branch’s (EGM’s) results to date indicate that the MUTSPEC 2.0 project can generate important knowledge on carcinogen properties. The obtained toxicogenomic signatures, when confirmed in collections of human samples, also have the potential to serve as biomarkers applicable to cancer etiology studies, early detection, and prevention measures.
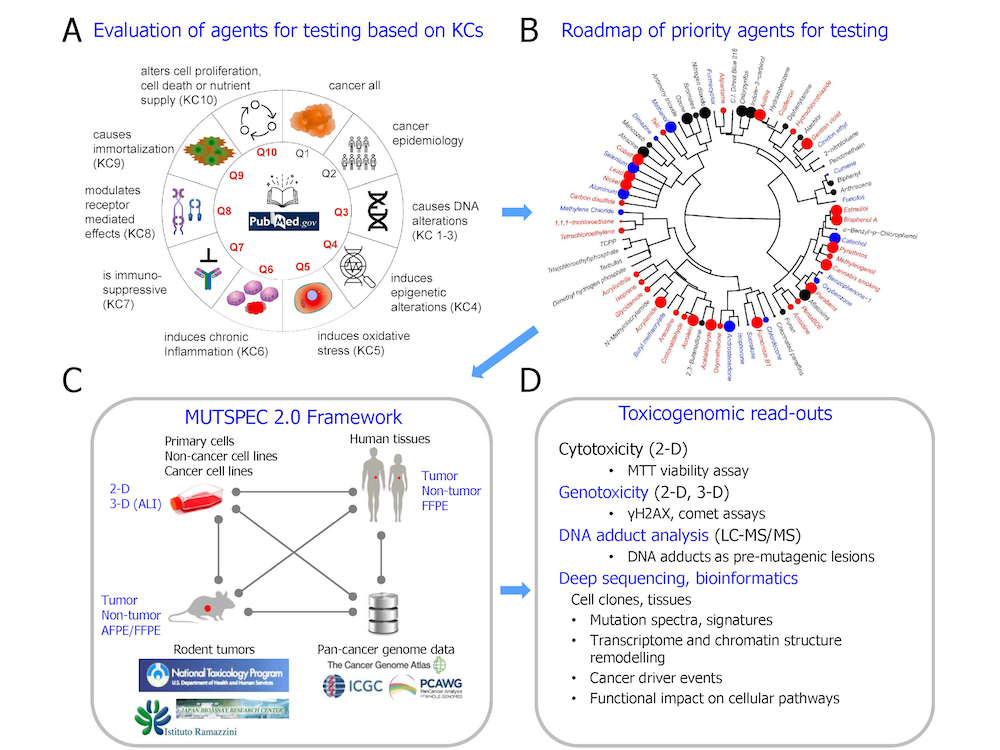
A schematic representation of the interdisciplinary, multi-system approach used in the MUTSPEC 2.0 project: (A) Candidate agents are selected based on key characteristics of carcinogens (KCs) used in prioritizing cancer hazard assessments; (B) A roadmap of candidate priority agents for testing is generated by chemoinformatics approaches (adapted from Barupal KD et al., Environ Int. 2021); (C) Candidate agents and their effects are evaluated in highly controlled in vitro and in vivo experimental exposure models, in human cancer samples, and by screening public cancer genome databases, within the framework of the MUTSPEC 2.0 project; (D) Examples of toxicogenomic read-outs and signatures obtained from the MUTSPEC 2.0 project. AFPE/FFPE, alcohol-fixed or formalin-fixed, paraffin-embedded (tissue); 2-D, 3-D, two-dimensional or three-dimensional cell cultures; ALI, air-liquid interface; ICGC, International Cancer Genome Consortium; LC-MS/MS, liquid chromatography–tandem mass spectrometry; PCAWG, Pan-Cancer Analysis of Whole Genomes.